Brain Tumor Segmentation with OpenCV, Python
₹5,000.00 Exc Tax
The project presents the MRI brain diagnosis support system for structure segmentation and its analysis using K-means clustering technique integrated with Fuzzy C-means algorithm. The method is proposed to segment normal tissues such as White Matter, Gray Matter, Cerebrospinal Fluid and abnormal tissue like tumour part from MR images automatically.
Platform : Python
Delivery Duration : 3-4 working Days
100 in stock
Description
ABSTRACT
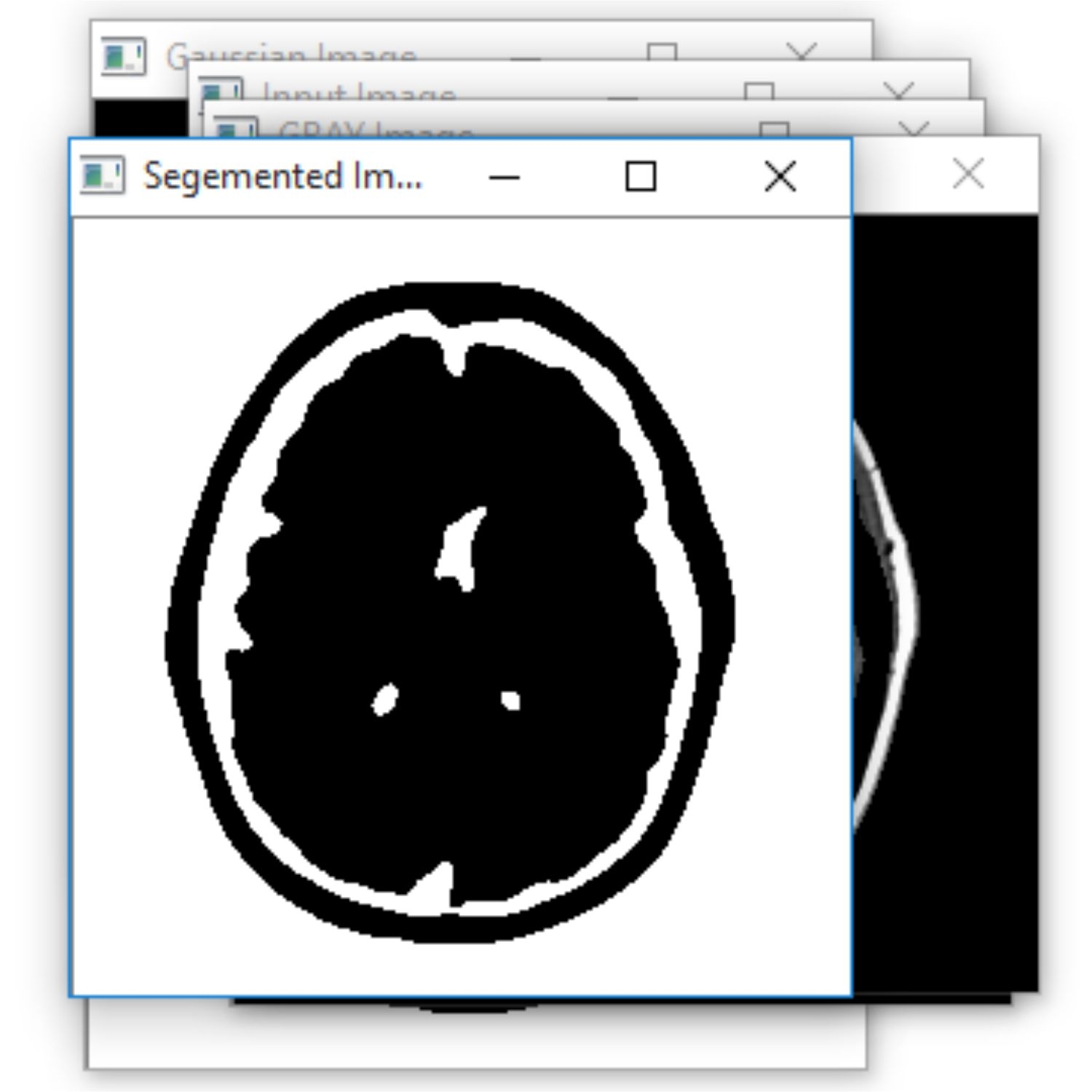
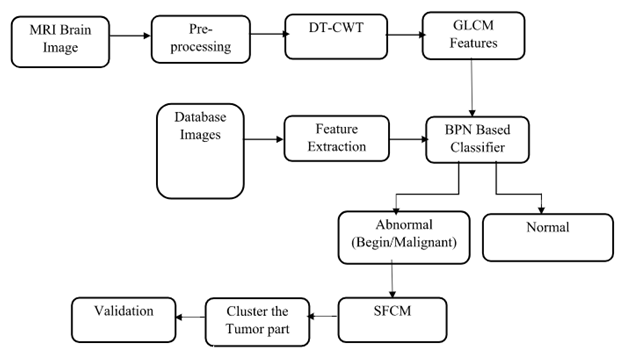
The project presents the MRI brain diagnosis support system for structure segmentation and its analysis using K-means clustering technique integrated with Fuzzy C-means algorithm. The method is proposed to segment normal tissues such as White Matter, Gray Matter, Cerebrospinal Fluid and abnormal tissue like tumour part from MR images automatically. These MR brain images are often corrupted with Intensity In homogeneity artefacts cause unwanted intensity variation due to non- uniformity in RF coils and noise due to thermal vibrations of electrons and ions and movement of objects during acquisition which may affect the performance of image processing techniques used for brain image analysis. Due to this type of artefacts and noises, sometimes one type of normal tissue in MRI may be misclassified as other type of normal tissue and it leads to error during diagnosis. The proposed method consists of pre-processing using Gaussian filter to remove noise and K-means clustering technique integrated with Fuzzy C-means algorithm segments normal tissues by considering spatial information because neighbouring pixels are highly correlated and also construct initial membership matrix randomly. The system also uses to segment the tumour cells along with this morphological filtering will be used to remove background noises for smoothening of region. The project results will be presented as segmented tissues and classification using neural network classifier
DEMO VIDEO
INTRODUCTION
In many recent object recognition systems, feature extraction stages are generally composed of a filter bank, a non-linear transformation, and some sort of feature pooling layer. Most systems use only one stage of feature extraction in which the filters are hard-wired, or two stages where the filters in one or both stages are learned in supervised or unsupervised mode. This paper addresses three questions: 1. How does the non-linearity’s that follow the filter banks influence the recognition accuracy? 2. does learning the filter banks in an unsupervised or supervised manner improve the performance over random filters or hardwired filters? 3. Is there any advantage to using an architecture with two stages of feature extraction, rather than one? We show that using non-linearity’s that include rectification and local contrast normalization is the single most important ingredient for good accuracy on object recognition benchmarks. We show that two stages of feature extraction yield better accuracy than one. Most surprisingly , we show that a two-stage system with random filters can yield almost 63% recognition rate on Caltech-101, provided that the proper non-linearities and pooling layers are used. Finally, we show that with supervised refinement, the system achieves state-of-the-art performance on NORB dataset (5.6%) and unsupervised pre-training followed by supervised refinement produces good accuracy on Caltech-101 (> 65%), and the lowest known error rate on the undistorted, unprocessed MNIST dataset (0.53%).
EXISTING SYSTEM
Earlier systems will detect the tumor but the drawback in the existing system is that the stages in the tumor identification is somewhat tedious process, without the level and stage of the tumor further proceeding to the treatment will not be desired.
PROPOSED SYSTEM
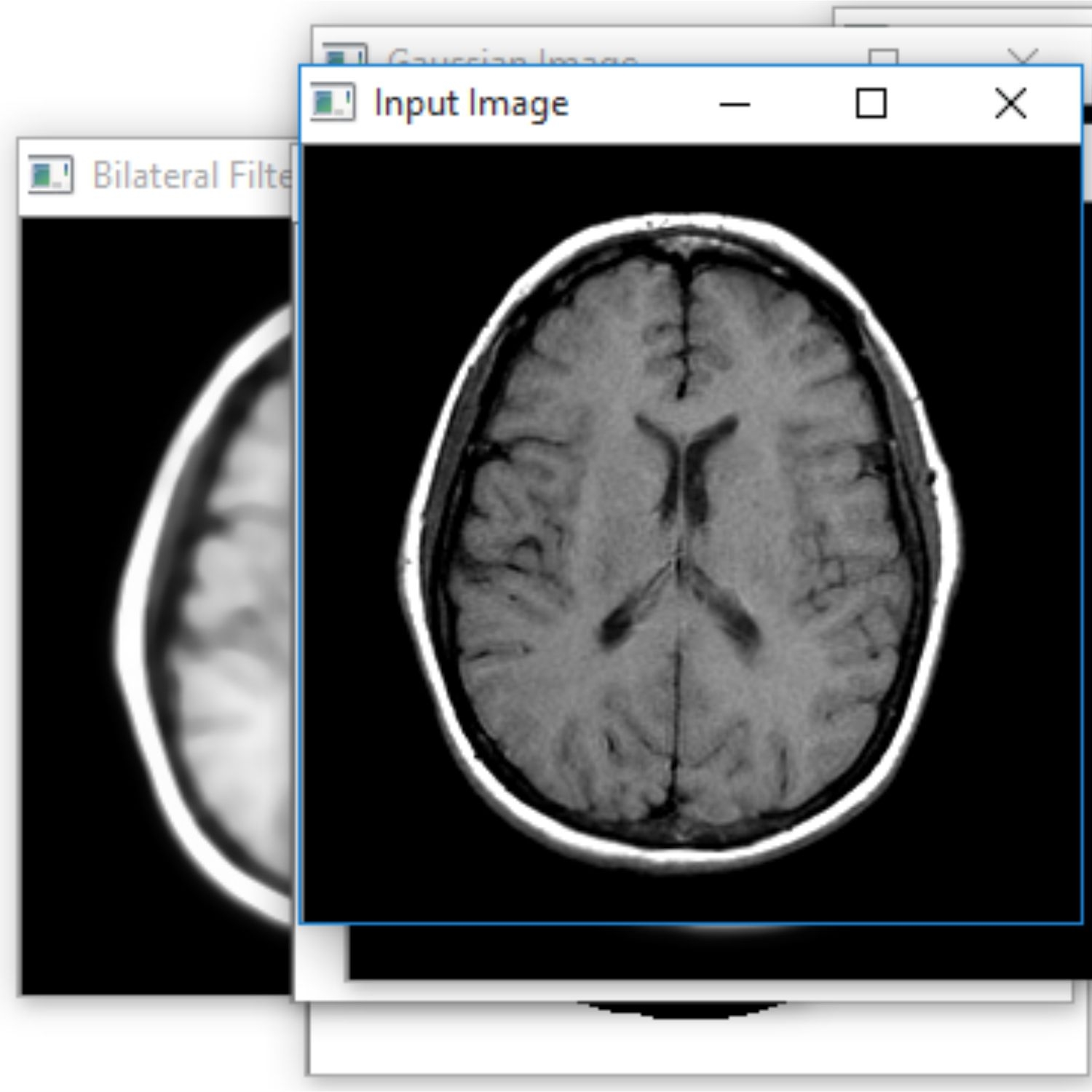
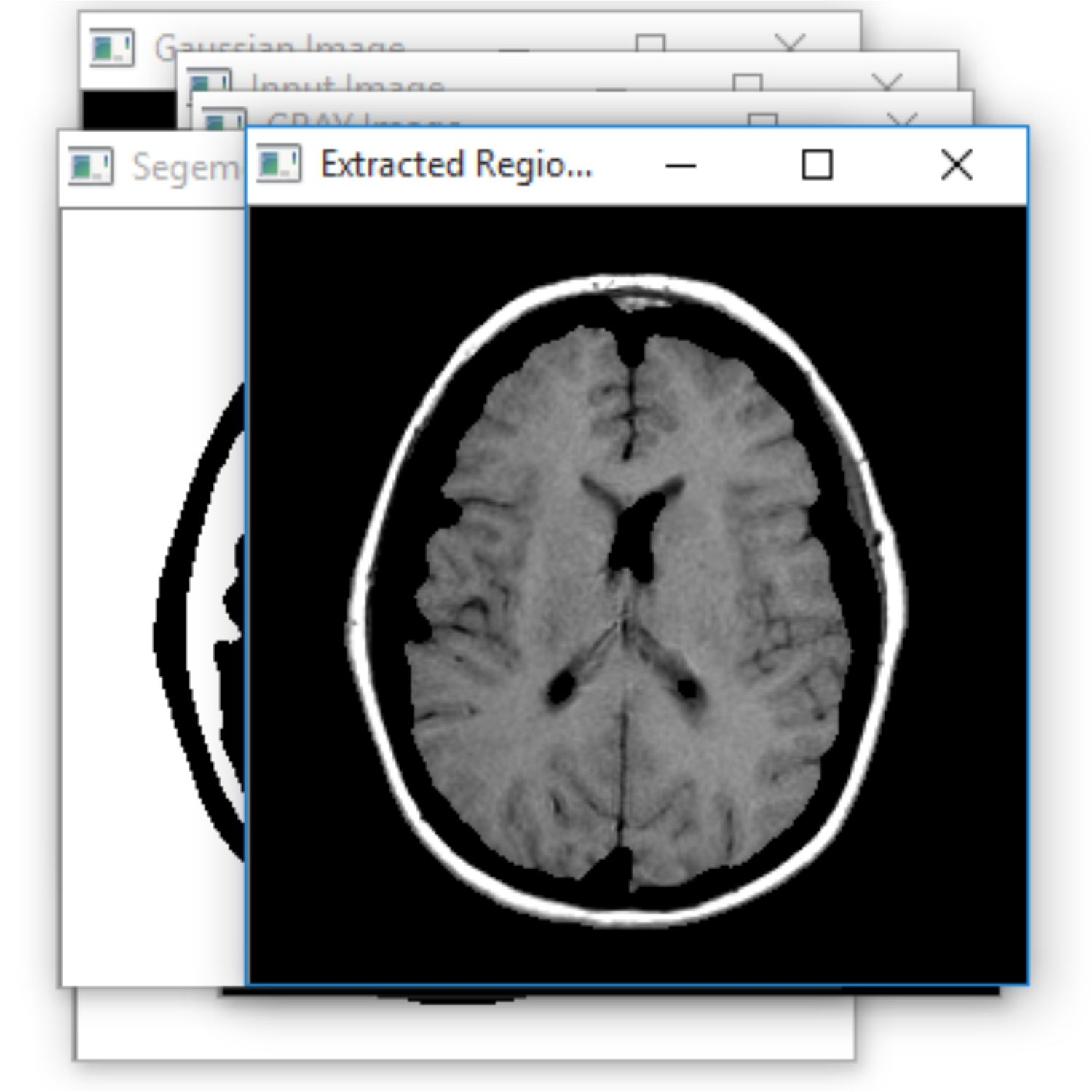
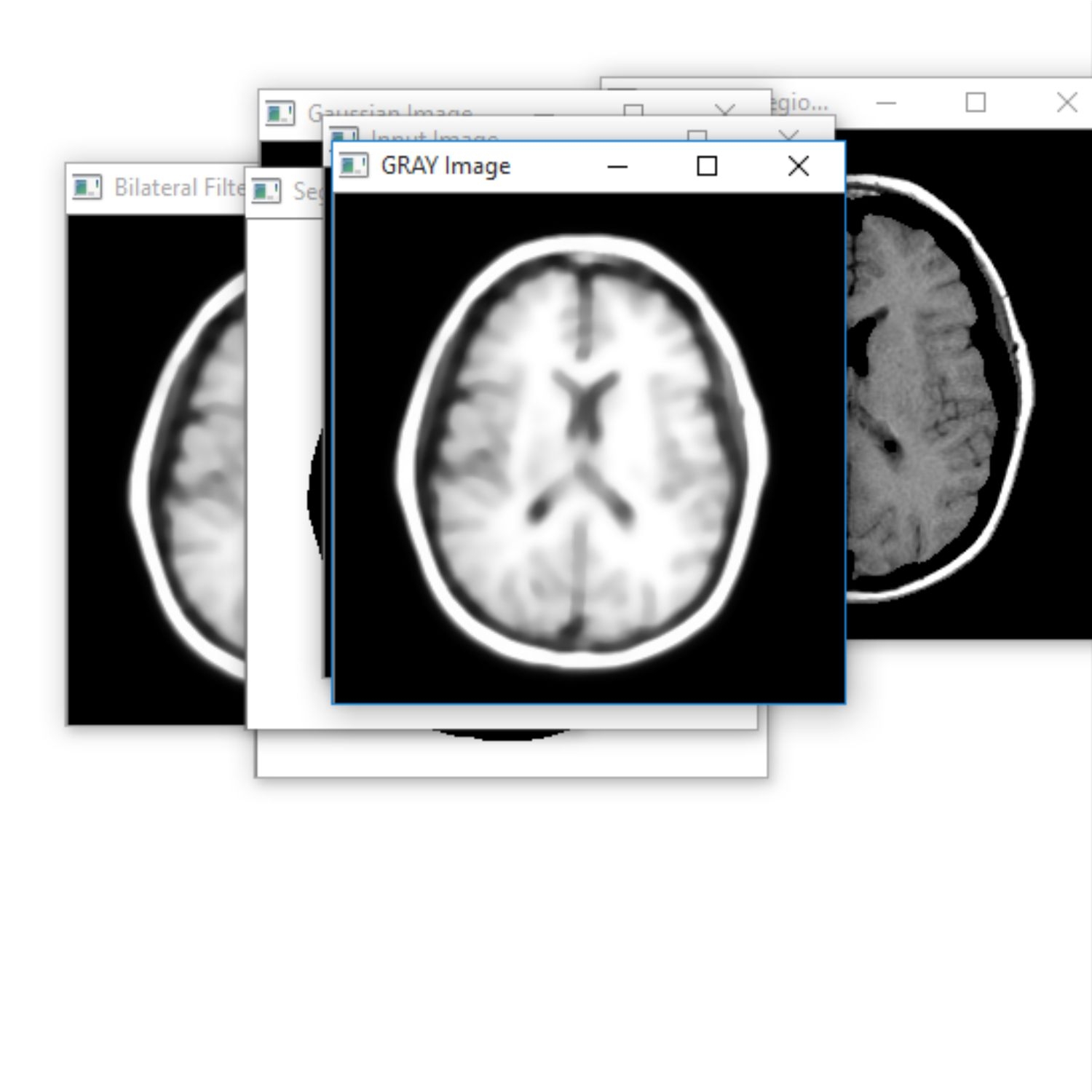
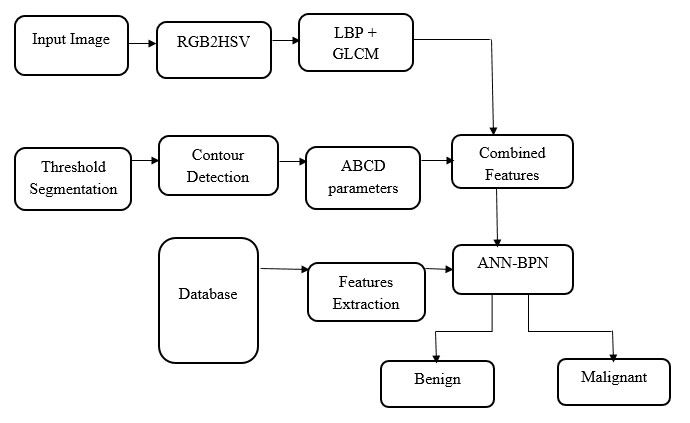
The Project proposes to spot the tumor from MRI scanned medical images using multi clustering model and morphological process. The segmentation refers to the process of partitioning a digital image into multiple segments. The brain MRI is taken and its noises are removed using filters and then applied spatial Fuzzy C means Clustering algorithm for the segmentation of MRI brain images. The morphological process will be used to smooth the tumor region from the noisy background. The segmented primary and secondary regions are compressed with hybrid techniques for telemedicine application.
NEURAL NETWORK
Neural networks are predictive models loosely based on the action of biological neurons. The selection of the name “neural network” was one of the great PR successes of the Twentieth Century. It certainly sounds more exciting than a technical description such as “A network of weighted, additive values with nonlinear transfer functions”. However, despite the name, neural networks are far from “thinking machines” or “artificial Lungs”. A typical artificial neural network might have a hundred neurons. In comparison, the human nervous system is believed to have about 3×1010 neurons. We are still light years from “Data”. The original “Perceptron” model was developed by Frank Rosenblatt in 1958. Rosenblatt’s model consisted of three layers, a “retina” that distributed inputs to the second layer, “association units” that combine the inputs with weights and trigger a threshold step function which feeds to the output layer, (3) the output layer which combines the values. Unfortunately, the use of a step function in the neurons made the perceptions difficult or impossible to train.
BLOCK DIAGRAM
ADVANTAGES
Using this system we can identify the different stages in the brain tumor, based on the stage of the tumor such as normal, moderate and severe, further treatment can be proceeded based on the results.
SOFTWARE REQUIREMENTS
- Python
- Opencv
- Numpy
RESULT
The proposed system is to identify the tumor with the use of neural networks. Based on that further treatment can be proceeded. Main advantage is that the identification of the level. It is the main in the clinical treatment.
REFERENCES
[1] Ardizzone.E, Pirrone.R, and Orazio.O.G, “Fuzzy C-Means Segmentation on Brain MR Slices Corrupted by RF-Inhomogeneity,” In Proc. The 7th international workshop on Fuzzy Logic and Applications: Applications of Fuzzy Sets Theory, WILF ’07, Springer- Verlag, pp: 378-384, 2007.
[2] Bianrgi.P.M, Ashtiyani.M, and Asadi.S, “MRI Segmentation Using Fuzzy C-means Clustering Algorithm Basis Neural Network,” In Proc. ICTT A 3rdInternationai Conference on Information and Communication Technologies: From Theory to Applications, pp: 1-5, 2008.
[3] Sikka.K, Sinha.N, Singh.P.K, and “A fully automated algorithm under modified FCM framework for improved brain MR image segmentation,” Magn. Reson. Imag, vol. 27, pp. 994–1004, Jul. 2009.
[4] Xiao.K, Ho.S.H, and Bargiela.B, “Automatic Brain MRI Segmentation Scheme Based on Feature Weighting Factors Selection on Fuzzy C means Clustering Aigorithms with Gaussian Smoothing,” International Journal of Computational Intelligence in Bioinforrnatics and Systems Biologyl(3):316-331,2009.
Additional information
| Weight | 0.000000 kg |
|---|



Reviews
There are no reviews yet.